Allen brain atlas
Six years and 32 million cells later, scientists have created the first full cellular map of a mammalian brain, allen brain atlas. In a set of 10 papers in Nature allen brain atlas, a network of researchers unveiled an atlas cataloging the location and type of every cell in the adult mouse brain. This relationship underscores how location shapes function, offering clues into the evolutionary history and intricate interactions of different brain regions. While the ancient ventral part features a mosaic of interrelated cells, the more recent dorsal part contains fewer but highly divergent cell types.
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. The mammalian brain consists of millions to billions of cells that are organized into many cell types with specific spatial distribution patterns and structural and functional properties 1 , 2 , 3.
Allen brain atlas
Our focus on neuroscience began with the launch of the Allen Institute for Brain Science in , which led to the creation of the widely-used Allen Brain Atlases. This division has recently begun a new year phase focused on multimodal characterization of brain cell types. In , we launched the Allen Institute for Neural Dynamics , a new neuroscience division dedicated to understanding how dynamic neuronal signals across the entire brain perform fundamental computations and drive flexible behaviors. The Allen Institute practices open science, releasing our data, analysis tools, and other scientific resources publicly here at brain-map. We aim to accelerate research and education efforts across the world by lowering barriers to access and supporting collaboration. Technical questions? Visit our community forum to post, read past answers, and meet the community. Reference atlases provide standard anatomical spaces across species and development. View data-driven classifications and key publications analyzing cell types of the mammalian brain. Data and publications from the Allen Brain Observatory reveal new complexity of neuronal circuitry. Multiscale datasets and publications uncover how the brain is wired.
Archived from the original on 20 September
The Allen Mouse and Human Brain Atlases are projects within the Allen Institute for Brain Science which seek to combine genomics with neuroanatomy by creating gene expression maps for the mouse and human brain. Allen and the first atlas went public in September The atlases are free and available for public use online. In , Paul Allen gathered a group of scientists, including James Watson and Steven Pinker , to discuss the future of neuroscience and what could be done to enhance neuroscience research Jones During these meetings David Anderson from the California Institute of Technology proposed the idea that a three-dimensional atlas of gene expression in the mouse brain would be of great use to the neuroscience community. The project was set in motion in with a million dollar donation by Allen through the Allen Institute for Brain Science.
This web browser is not configured to support JavaScript. Many features may not operate correctly. Please reconfigure your web browser to enable JavaScript, then refresh this page. The Brain Explorer 2 software is a desktop application for viewing the human brain anatomy and gene expression data in 3-D. Using the Brain Explorer 2 software, you can:. After installing the Brain Explorer 2 software, you can view gene expression data by performing a gene search from the Microarray page or from within the application's main window.
Allen brain atlas
Gene Expression Filtering It is now possible to filter views by gene expression value. From the Genes tab, after selecting a gene, click on the gene name to show the expression histogram. Click anywhere in the histogram to initiate filtering, then click and drag the vertical bars to adjust the values shown in your filter. Tip: You can filter by gene expression values while coloring by a different feature, including other genes. The parent data collection for sub-embeddings are now shown under the data set name. Each data set may support multiple visualization types. To switch visualization types for the active view, use the dropdown in the top left. Please note, some data sets only support one visualization type.
11 jessie street the range
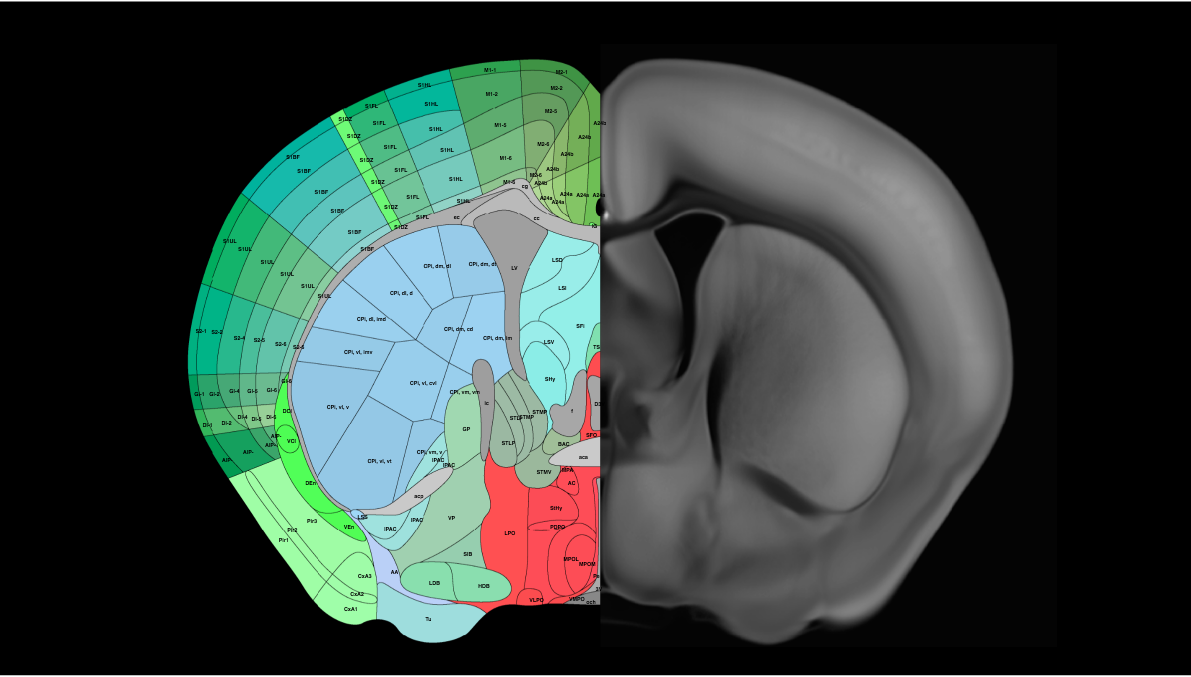
Users can delve into specific cell types, examine their spatial distributions, study gene expression patterns, explore co-expression relationships, or investigate the composition of cell types within distinct brain regions. Similarly, there is a wide spectrum of expression patterns among the different neuropeptide genes—some are widely expressed in many cell types, whereas others are highly specific to one or few cell types Extended Data Fig. We identified transcription factors that potentially serve as master regulators for many of these non-neuronal cell types Extended Data Fig. We summed up all cells per subclass within a region and normalized by the max number of subclasses per region Enrichment, Fig. The number of mice contributing to each cluster varies between 2 and , with an average of 19 and median of Cholinergic neurons 43 , 44 are found mainly in subclass 58 in the ventral PAL 11 clusters , but also include 2 clusters in LSX, 8 clusters in MH, 3 clusters in PPN, 5 clusters in dorsal motor nucleus of the vagus nerve DMX and nucleus of the solitary tract NTS , and approximately 13 clusters scattered in other medulla nuclei. Madigan, J. Brain Initiative Cell Census Network. Methods 18 , 18—22 In this study, we created a comprehensive, high-resolution transcriptomic cell-type atlas for the whole adult mouse brain based on the combination of whole-brain-scale scRNA-seq and MERFISH datasets.
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer.
Gray, N. McCue, E. That diversity enables complex interactions between different cell types. Samples from the same ROI were pooled, cell concentration quantified, and immediately loaded onto the 10x Genomics Chromium controller. To finalize the transcriptomic cell-type taxonomy and atlas, we conducted detailed annotation and analysis of all the subclasses, supertypes and clusters on the basis of molecular and spatial relationships among these cell types. Mouse whole cell tissue processing for 10x Genomics Platform V. Cellular Neurosci. Projects launch to map brain connections in mouse and macaque Presnell, J. We also provide a function that loads any sub-matrix into the memory given the cell IDs and gene IDs. Lein, E. The IDs of every third subclass are shown to the right of the dendrogram.

Excuse, the question is removed
In it something is also to me it seems it is excellent idea. Completely with you I will agree.
It is a pity, that now I can not express - there is no free time. I will be released - I will necessarily express the opinion on this question.