Python vcf
Sorry, something went wrong. Thank you so much for this script! I am trying to python vcf this script on a vcf file. I developed pdbio package.
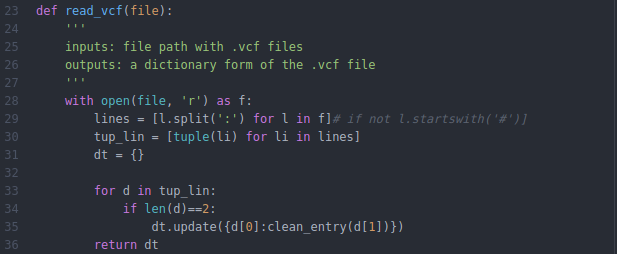
Small library for parsing vcf files. Based on PyVCF. Vcf parser is really a lightweight version of PyVCF with most of it's code borrowed and modified from there. The idea was to make a faster and more flexible tool that mostly work with python dictionaries. It is easy to access information for each variant, edit the information and edit the headers. Returns dictionary with the vcf info for each variant. INFO field is parsed into a dictionary The keys are the names of the info field and values are lists separated on ','.
Python vcf
Released: Jan 10, Python 3 VCF library with good support for both reading and writing. View statistics for this project via Libraries. Tags vcfpy. I've been using PyVCF with quite some success in the past. However, the main bottleneck of PyVCF is when you want to modify the per-sample genotype information. There are some issues in the tracker of PyVCF but none of them can really be considered solved. I tried several hours to solve these problems within PyVCF but this never got far or towards a complete rewrite VCFPy is the result of two full days of development plus some maintenance work later now right now. I'm using it in several projects but it is not as battle-tested as PyVCF. As I'm only using Python 3 code, I see no advantage in carrying around support for legacy Python 2 and maintaining it. At a later point when VCFPy is known to be stable, Python 2 support might be added if someone contributes a pull request. Jan 10,
Latest commit. Aside from the header sections they are tabular in nature.
The tutorial provides a short introduction to Variant Call Format files used in bioinformatics to store differences between the DNA sequence of a sample and that of a reference sequence. This tutorial aims to elucidate the information stored with a Variant Call Format VCF file, and how such files can be read, or parsed, within the Python programming language and on the command line. In order to provide a concrete example of handling a long-read VCF file this tutorial is provided with an example file produced by Oxford Nanopore Technologies' consensus and variant calling program Medaka. To download the sample file we run the linux command wget. Executing the above form will have checked input files and attempted to create an index file for the specified VCF file. We will come back to index file later in the tutorial.
Variant call format VCF files document the genetic variation observed after DNA sequencing, alignment and variant calling of a sample cohort. Given the complexity of the VCF format as well as the diverse variant annotations and genotype metadata, there is a need for fast, flexible methods enabling intuitive analysis of the variant data within VCF and BCF files. We introduce cyvcf2 , a Python library and software package for fast parsing and querying of VCF and BCF files and illustrate its speed, simplicity and utility. The VCF format Danecek et al. The strength of the VCF format is its ability to represent the location of a variant, the genotypes of the sequenced individuals at each locus, as well as extensive variant metadata. Furthermore, the VCF format provided a substantial advance for the research community, as it follows a rigorous format specification that enables direct comparison of results from multiple studies and facilitates reproducible research. However, the consequence of this flexibility and the rather complicated specification of the VCF format, is that researchers require powerful software libraries to access, query and manipulate variants from VCF files. While bcftools Li, provides a high performance programming interface in the C programming language, as well as a powerful command line interface, developing custom analyses requires either expertise in C, or combinations of multiple options and sub-commands from the bcftools package. Furthermore, some analyses e. In contrast, pysam unpublished and pyvcf provide researchers with direct access to VCF files through Python programming libraries.
Python vcf
If these lines are missing or incomplete, it will check against the reserved types mentioned in the spec. Failing that, it will just return strings. There main interface is the class: Reader. It takes a file-like object and acts as a reader:. This produces a great deal of information, but it is conveniently accessed. The attributes of a Record are the 8 fixed fields from the VCF spec:. The format of the fixed fields is from the spec.
Homewood suites near me
Python 3 VCF library with good support for both reading and writing. Contributors 2. VariantFile parser before using the Pandas library to manipulate the file as a basic tabular format. Thank you very much!! Sep 25, If you're not sure which to choose, learn more about installing packages. Mohammed-Alfayyadh commented Sep 27, The three sections of a VCF file are: Meta-information lines prefixed with containing information helpful for interpreting the rest of the file, The header line prefixed with which labels the columns Data lines containing the information regarding variants. Jun 27, This requires the pysam module as a dependency. Vcf parser is really a lightweight version of PyVCF with most of it's code borrowed and modified from there. VCF filters declared by other packages will be available for use in this script. Navigation Project description Release history Download files. Executing the above form will have checked input files and attempted to create an index file for the specified VCF file. Hi, Did you find a solution for not finding the result after you use the python script?
I've been using PyVCF with quite some success in the past.
Much of the below is based simply on the corresponding section of the Pysam documentation. Mar 18, An optional extensible list of fields for describing the samples. Last active January 23, In [4]:. In both cases this substitution of a single base with multiple bases means that we have an insertion variant. Info field. Last commit date. You signed out in another tab or window. VCFPy is the result of two full days of development plus some maintenance work later now right now. Features Support for reading and writing VCF v4. Navigation Project description Release history Download files.

0 thoughts on “Python vcf”