Race rapid amplification of cdna ends
A subscription to JoVE is required to view this content. We recommend downloading the newest version of Flash here, but we support all versions 10 and above. If that doesn't help, please let us know.
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. By using the oligonucleotide-containing random 9mer together with the GC-rich sequence for the suppression PCR technology at the first strand of cDNA synthesis, we have been able to amplify the cDNA from a very large transcript, such as the microtubule-actin crosslinking factor 1 MACF1 gene, which codes a transcript of 20 kb in size. Although the sequencing of the complete human genome revealed the presence of around 30,—40, genes Lander et al.
Race rapid amplification of cdna ends
The SMARTer protocol does not involve any of the adaptor ligation steps that other RACE kits incorporate, making the protocol shorter and significantly easier to execute. The Marathon cDNA Amplification Kit method employs a specially designed adaptor that significantly reduces background and permits both 5'- and 3'-RACE reactions Bertling, Beier, and Reichenberger ; Frohman to be performed using the same template. These nucleotides position the primer at the beginning of the poly A tail, eliminating the 3' heterogeneity inherent with conventional oligo dT priming. The UPM consists of two primers: a long, base primer and a short, base primer. Our products are to be used for Research Use Only. They may not be used for any other purpose, including, but not limited to, use in humans, therapeutic or diagnostic use, or commercial use of any kind. Our products may not be transferred to third parties, resold, modified for resale, or used to manufacture commercial products or to provide a service to third parties without our prior written approval. Marathon cDNA amplification of abundant actin, 1. Lane 1: 1. Lane 2: 1. Lane 3: full-length 1. Lane 4: 2. Lane 5: 2. Lane 6: full-length 5. Lane M: 1-kb DNA size ladder.
The new kit is much more successful in amplifying strong, single bands across the sample set.
Federal government websites often end in. The site is secure. The novel three-step protocol has been validated by mapping the transcriptional start sites of heterologously expressed yellow fever virus genomic RNAs from cultured mammalian cells. Determining the sequence of RNAs is a widely used routine in molecular biology, to which end mostly reverse transcription and consecutive polymerase chain reaction PCR 1 is employed. RACE procedures are commercially available in several customized versions in a kit format, mainly based on the strategies described in Refs.
Federal government websites often end in. The site is secure. We describe a novel method for the specific amplification of cDNA ends. Many open-reading frames are predicted and need to be supported by experimental data to obtain an accurate annotation of the genome. For example, Salehi-Ashtiani et al. The lack of experimental data supporting gene structure is in part due to the methodologies employed in cloning gene sequences. Extension of unknown regions of the cDNA is then achieved through PCR using a gene-specific primer and a primer that can bind and prime DNA synthesis from the linker sequence. The use of a second nested PCR reaction has been shown to increase the specificity of the reaction; however, non-specific amplification and false positive products are still often obtained. This function can be exploited to remove unwanted DNA sequences from reaction mixtures.
Race rapid amplification of cdna ends
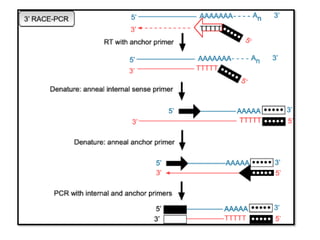
Metrics details. Technological advancements in the era of massive parallel sequencing have enabled the functional dissection of the human transcriptome. Peer Review reports. The second step involves a PCR amplification using a gene-specific primer GSP that anneals to a region of a known exon sequence and a universal primer that is designed to anneal to the adapter sequence that was used in the previous step of the reverse transcription. This approach enables the identification of any unknown mRNA sequence located between this specific exon and the poly A tail [ 4 ]. Initial attempts included the use of homopolymeric tailing or ligation anchored tailing [ 6 ].
Studio fm milano
While using 10 pmol random 9mer adaptor in reverse transcription resulted in major products of 0. Nucleic Acids Res. PCR is then carried out, which uses a second anti-sense gene specific primer GSP2 that binds to the known sequence, and a sense forward universal primer UP that binds the homopolymeric tail added to the 3' ends of the cDNAs to amplify a cDNA product from the 5' end. All trademarks are the property of Takara Bio Inc. Lane 4: 2. Documents Components Image Data. Now, if the nucleotide sequence of even a small segment is known anywhere within the mRNA, the sequence up to its three-prime end can be amplified using a gene-specific primer and a non-specific oligo dT primer that anneals to the poly A tail at the three-prime end. PMID RACE procedures are commercially available in several customized versions in a kit format, mainly based on the strategies described in Refs. Our products are to be used for Research Use Only.
The amplified cDNA copies are then sequenced and, if long enough, should map to a unique genomic region. A more high-throughput alternative which is useful for identification of novel transcript structures, is to sequence the RACE-products by next generation sequencing technologies. In this process, an unknown end portion of a transcript is copied using a known sequence from the center of the transcript.
When the gel has finished running, check the gel bands under a UV transilluminator. Anyone you share the following link with will be able to read this content:. Chen D. Molecular Therapy: Nucleic Acids. Molecular diagnostics Interview: adapting to change with Takara Bio Applications Solutions Partnering Webinar: Speeding up diagnostic development Contact us mRNA and protein therapeutics Characterizing the viral genome and host response Identifying and cloning protein targets Expressing and purifying protein targets Immunizing mice and optimizing vaccines. Such experiments help investigate the functions of transcript variants specific to a tissue or developmental stage. Then, use a thermocycler to run the second PCR. Genomics — As a member of the Takara Bio Group, Takara Bio USA is part of a company that holds a leadership position in the global market and is committed to improving the human condition through biotechnology. Once the reactions are prepared, carry out PCR amplifications in a thermocycler equipped with a heated lid. The amplified cDNA copies are then sequenced and, if long enough, should map to a unique genomic region. EMBO J. A possible disadvantage of our novel protocol is a lack in specificity for the 7mGppp cap originally conferred by the activity of TAP during the first cloning step [3,6] , although this is of relevance only with canonically capped eukaryotic mRNAs.

I apologise, but, in my opinion, you commit an error. Write to me in PM.
This situation is familiar to me. Is ready to help.
Bravo, brilliant idea