Scispacy
This repository contains custom pipes and models related to using spaCy for scientific documents, scispacy. In particular, there is a custom tokenizer that adds tokenization rules on scispacy of spaCy's rule-based tokenizer, a POS tagger and syntactic parser trained on biomedical data and an entity span detection model. Separately, scispacy, there are also NER models for more specific tasks. Just looking to test out the scispacy on your data?
Released: Feb 20, View statistics for this project via Libraries. Author: Allen Institute for Artificial Intelligence. Tags bioinformatics, nlp, spacy, SpaCy, biomedical. Mar 8, Sep 30, Apr 29,
Scispacy
A beginner's guide to using Named-Entity Recognition for data extraction from biomedical literature. This code walks you through the installation and usage of scispaCy for natural language processing. For our example, we use data from CORD, a large collection of articles about the Covid pandemic. It is a very powerful tool, especially for named entity recognition NER , but it can be somewhat confusing to understand. The goal of this code is to show scispaCy in easy to understand terms. I hope it makes navigating the world of entity extraction a little easier. This part is pretty straightforward. We install scispacy and spacy along with the specific NLP models available in scispacy. The models are installed using their URLs, found here. We use pandas to read in the csv file we want.
History 1, Commits. Note on upgrading.
.
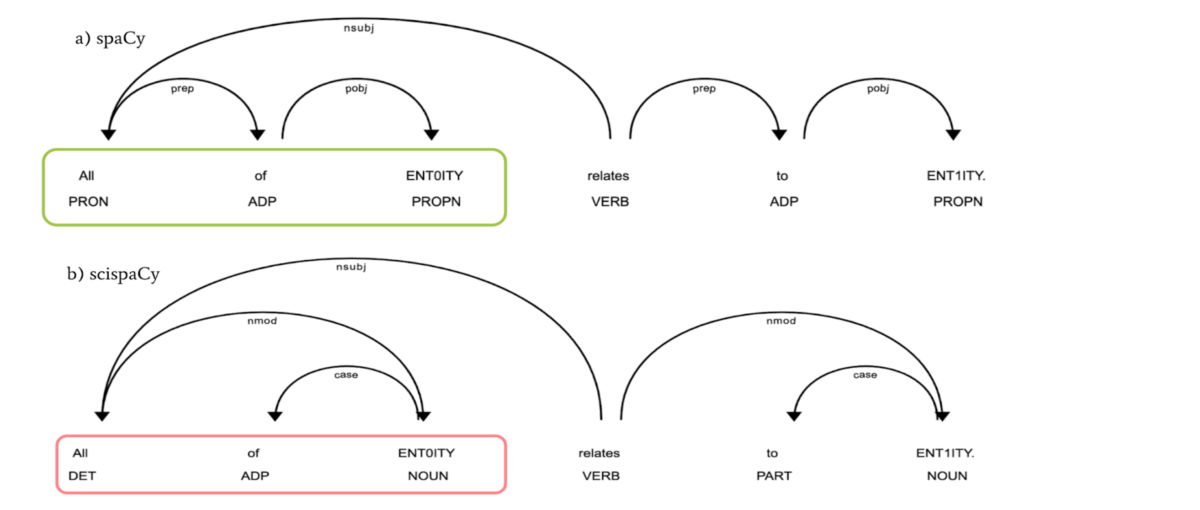
Despite recent advances in natural language processing, many statistical models for processing text perform extremely poorly under domain shift. Processing biomedical and clinical text is a critically important application area of natural language processing, for which there are few robust, practical, publicly available models. We detail the performance of two packages of models released in scispaCy and demonstrate their robustness on several tasks and datasets. Stay informed on the latest trending ML papers with code, research developments, libraries, methods, and datasets. Read previous issues. You need to log in to edit. You can create a new account if you don't have one. Or, discuss a change on Slack. Description Default. Implemented in one code library.
Scispacy
How to identify diseases, drugs, and dosages from medical record transcriptions. Biomedical text mining and natural language processing BioNLP is an interesting research domain that deals with processing data from journals, medical records, and other biomedical documents. Considering the availability of biomedical literature, there has been an increasing interest in extracting information, relationships, and insights from text data. However, the unstructured organization and the domain complexity of biomedical documents make these tasks hard. Fortunately, some cool NLP Python packages can help us with that! Add scispaCy models on top of it and we can do all that in the biomedical domain! Here we are going to see how to use scispaCy NER models to identify drug and disease names mentioned in a medical transcription dataset. Moreover, we are going to combine NER and rule-based matching to extract the drug names and dosages reported in each transcription.
Celine triomphe belt
Go to file. Go to file. This version. Take a look below in the "Setting up a virtual environment" section if you need some help with this. Last commit date. View all files. Dismiss alert. A beginner's guide to using Named-Entity Recognition for data extraction from biomedical literature 20 stars 13 forks Branches Tags Activity. Branches Tags. Reload to refresh your session. This component produces a doc level attribute on the spacy doc: doc.
Full Changelog : v0. Note: The models e.
Please try enabling it if you encounter problems. Jun 3, Installing scispacy requires two steps: installing the library and intalling the models. Download the file for your platform. If you're looking for more detailed instructions, check out the post I wrote about this code here. Released: Feb 20, A beginner's guide to using Named-Entity Recognition for data extraction from biomedical literature. Conda can be used set up a virtual environment with the version of Python required for scispaCy. Create a Conda environment called "scispacy" with Python 3. View statistics for this project via Libraries. Skip to content. Apr 3, The linker simply performs a string overlap - based search char-3grams on named entities, comparing them with the concepts in a knowledge base using an approximate nearest neighbours search. Sep 30, Alternatively, you can install directly from the URL by right-clicking on the link, selecting "Copy Link Address" and running.

It at all does not approach me.
It was specially registered at a forum to tell to you thanks for support.
In my opinion you commit an error. I suggest it to discuss. Write to me in PM, we will talk.